
Discover the latest advancements in muscle biology and exercise physiology.

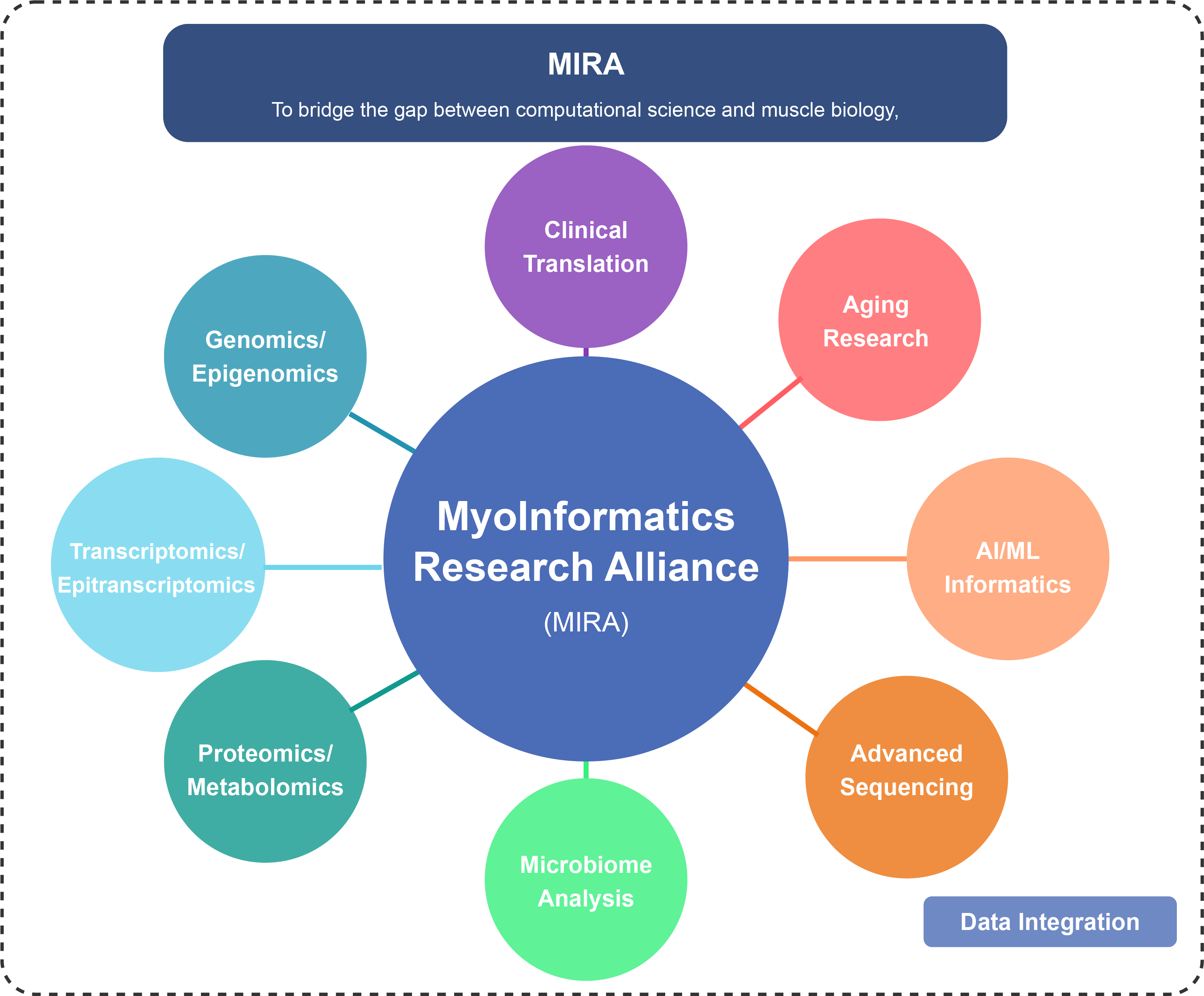
MIRA represents a collaborative research initiative focused on applying informatics and AI approaches to the study of skeletal muscle biology and related health conditions.
To bridge the gap between computational science and muscle biology, creating a future where comprehensive data integration and AI-driven analysis reveal previously hidden insights that advance human health and extend healthy muscle function throughout life.
MIRA's mission is to develop and implement advanced computational methods, multi-omics technologies, and AI-driven approaches that transform our understanding of muscle biology.
We accomplish this by:
Our alliance focuses on muscle biology and exercise physiology in health and disease, utilizing bioinformatics and omics technologies to understand the molecular mechanisms underlying muscle function and adaptation.
MIRA is developing in-house sequencing capabilities that combine both short-read and long-read technologies:
MIRA utilizes a comprehensive suite of sequencing techniques that span different levels of biological regulation:
MIRA is actively developing and optimizing novel sequencing protocols:
For more information, please reach out to us at ywen2@uky.edu.